Stanford-CIDE Coronavirus Simulation Model
What it does
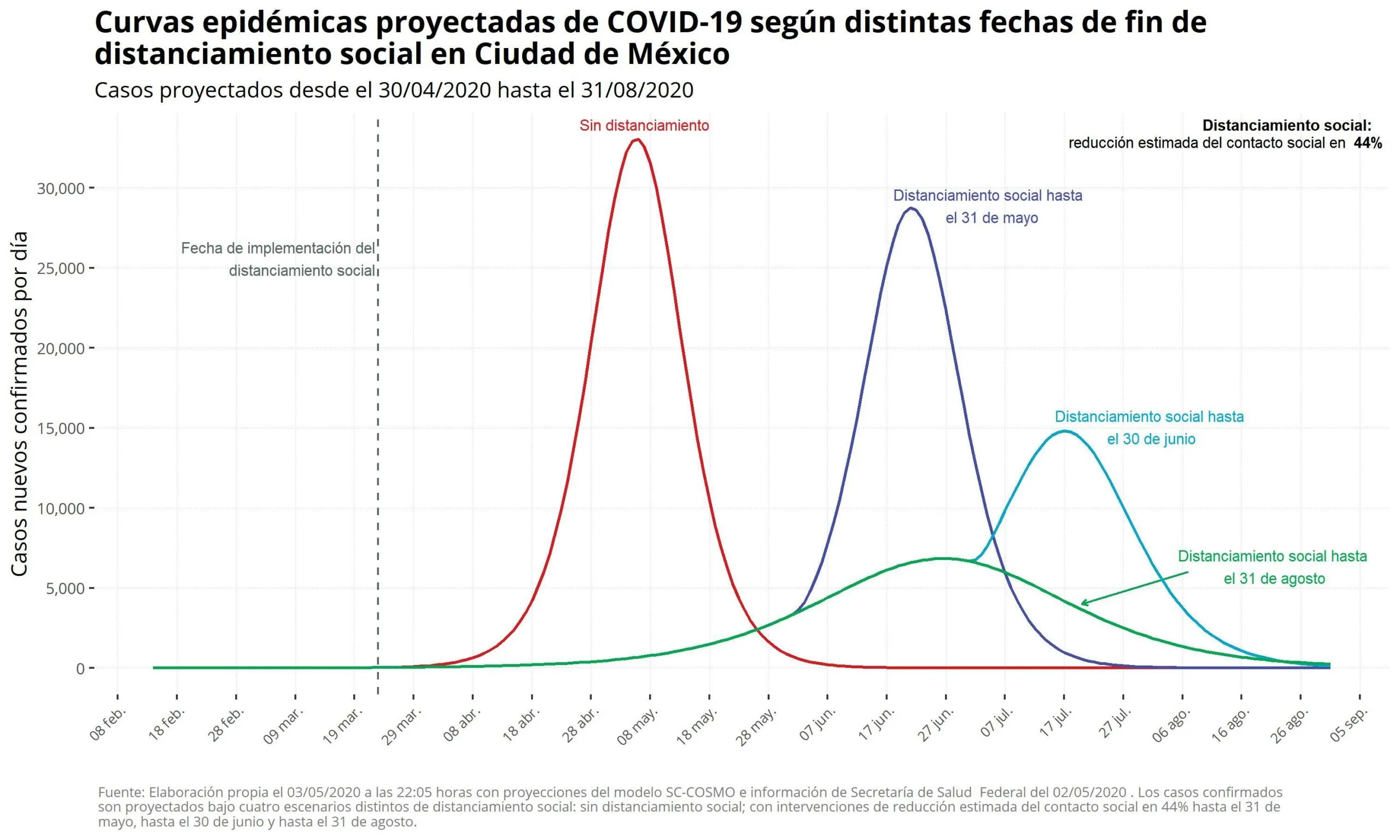
The SC-COSMO modeling framework enables modeling of the epidemiology of COVID-19 for diverse populations and geographies. At its core, it is an age-structured, multi-compartment susceptible-exposed-infected-recovered (AS-MC-SEIR) model implemented in the R programming language. The model incorporates realistic demography and patterns of contacts sufficient for transmission. The model also incorporates non-pharmaceutical interventions (NPIs) (e.g., “social distancing”) timing and effects on reductions in contacts which may differ by demography. The model framework also allows for the comparison of many, future what-if scenarios and how they might impact outcomes over time and cumulatively (e.g., infections, cases, deaths, etc.). The framework can be implemented rapidly for specific populations and geographies based on generally available data from these places along with methods we have implemented to infer data elements that are unavailable (e.g., imputation and calibration).
Why I made it
During the COVID-19 lockdown, I found myself with unexpected free time and a strong desire to put my data analysis, visualization, and research skills to meaningful use. I reached out to my undergraduate thesis advisor—who was leading the SC-COSMO project—and volunteered to contribute. This was both an opportunity to support a critical public health effort and a way to challenge myself technically during an uncertain time.
Tools & Technologies
- R – for implementing the AS-MC-SEIR simulation model
- RMarkdown – for documentation and reproducible reports
- ggplot2 / plotly – for data visualization
- Web scrapping - for extracting data from publicly available reports
- Parallel computing (e.g., doParallel, foreach) – to speed up calibration and scenario simulations
- Git – for version control and collaborative development
- Public COVID-19 datasets – for calibration (e.g., case counts, testing data, demographic data)
- Empirical contact matrices – for parameter inference across geographies
What I learned
Key takeaways include:
- Gained hands-on experience implementing and adapting a complex epidemiological model (AS-MC-SEIR) in R.
- Improved my ability to work with real-world data sources, including demographic and COVID-19 surveillance data.
- Learned to calibrate large models using noisy and incomplete datasets.
- Developed stronger skills in data visualization and communicating model outputs under uncertainty.
- Gained experience in optimizing code for speed using parallel processing in R.
Real-world use / impact
The SC-COSMO model played a key role in supporting real-time policy decisions during the COVID-19 pandemic in Mexico and Latin America. It provided government agencies and public health institutions with data-driven forecasts to evaluate the potential effects of non-pharmaceutical interventions like lockdowns, school closures, and mobility restrictions. By simulating dozens of “what-if” scenarios calibrated to local data, the model helped inform strategic decisions around reopening timelines and resource allocation. My contributions supported the generation of visual outputs and scenario analyses that were presented to health authorities, helping translate complex epidemiological dynamics into actionable insights at a time of great urgency.
Team
SC-Cosmo Consortium
🔗 Code
View on GitHub